Afra Shamnath
01/12/2021
First spotted in sequences deposited from Botswana, South Africa, multiple genomes of Omicron variant are now being sequenced from a number of countries around the world. The variant is characterised with 32 spike mutations. Initial reports suggest that the variant could be associated with immune escape and higher transmissibility. PCR assays and genome sequencing has aided in detection of the variant. The variant has now been detected in Hong Kong, Israel, Belgium, UK, Germany, Italy Czech Republic and anticipated in many other countries.
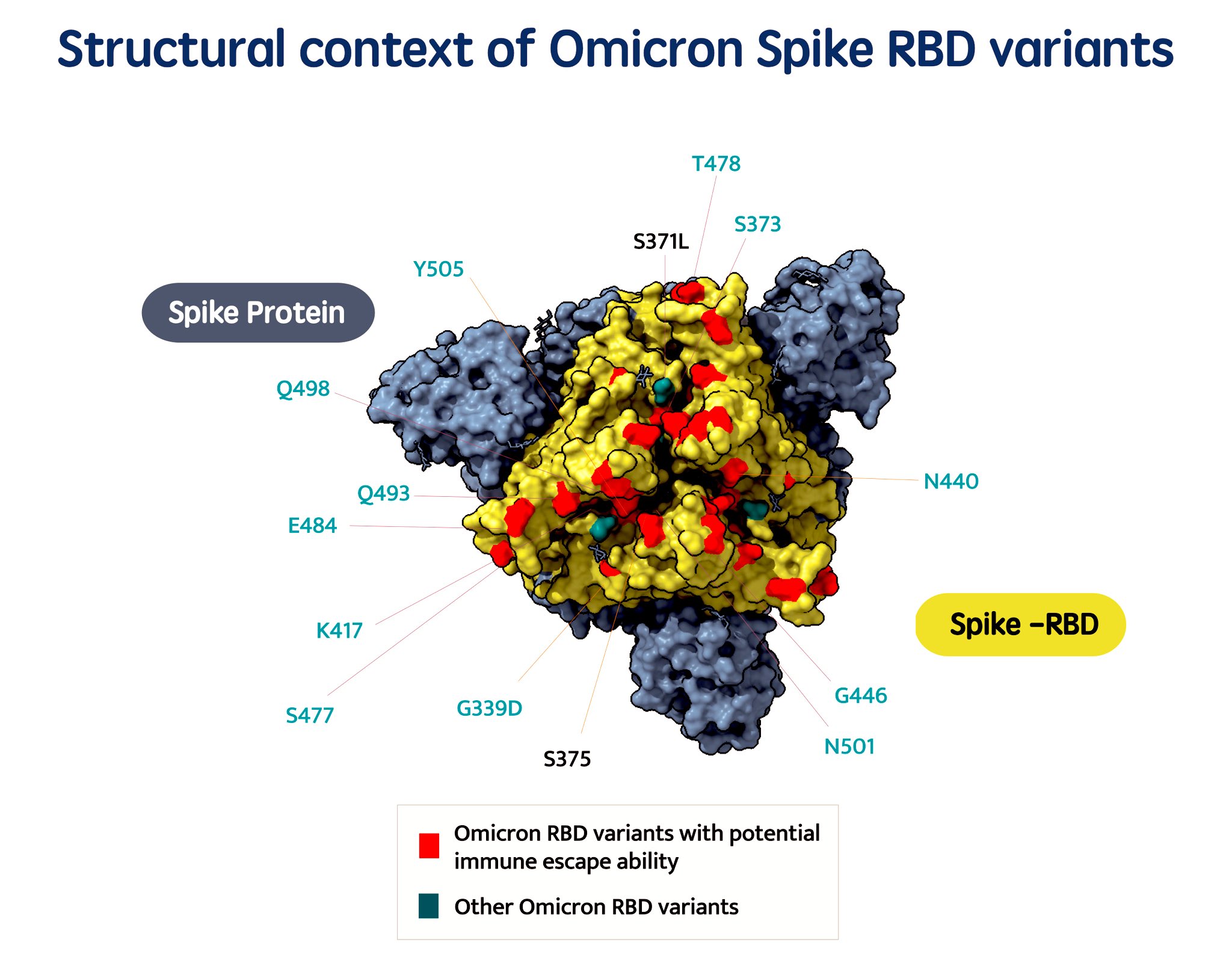
The initial sequences came from specific outbreaks in South Africa, thanks to the committed researchers in the country who were doggedly tracking outbreaks in the country and faithfully depositing the data in GISAID, a platform for open sharing of genomic data from outbreaks. Dr Tom Peacock, virologist at Imperial College who first reported the variant in PANGO, an open network of researchers analysing clusters of SARS-CoV-2 from genomic datasets took to Twitter to describe the “horrific profile” with 32 spike mutations. Mutation profiling of the variant has been unusually different from other variants of concern and has revealed infamous mutations in the Receptor Binding Domain(RBD) and N Terminal Domain(NTD). This could result in catastrophic consequences due to resistance of neutralising antibodies.
Jesse Bloom (@jbloom_lab), affiliated with Fred Hutch provided a wonderful preliminary insight on the how the variant will affect polyclonal and monoclonal antibodies targeting the Receptor Binding Domain(RBD) and they affirm that the variant has a lot of antigenic change. This points out to the fact that even in a polyclonal mix, the receptor binding domains will be affected.
The stats encapsulated by John Burn Murdoch, data journalist does raise an alarm or two. He says “There is a clear upward trend. This may be a blip, but this is how waves start.”He believes targeted sequencing and testing should be top priority. Dr Eric Topol, physician-scientist to CNN Live asks a valid query” The big question now is will our immune response from vaccines or prior infections be blunted by this variant? That isn't clear yet.” Dr Ben Cowling an epidemiologist at the University of Hong Kong says that the discovery of two imported cases of variant in Hong Kong suggests that the variant is already in international circulation. Tom Wenseleers, a statistician at KU Leuven suggests a 6 fold higher R value than Delta.
Mercy Rophina, graduate student at CSIR-IGIB using her Database of Immune Escape Variants in SARS-CoV-2 (ESC) analysed the functional implications and immune evasion capability of the lineage defining mutations revealed that about 23 variants possess proven functional consequences. In addition, 17 unique spike mutations were found to have immune escape mechanisms. It was interesting to find spike mutations with potential escape against mABs including LY-CoV016/Etesevimab, LY-CoV555/Bamlanivimab, REGN10933/Casirivimab, REGN10987/Imdevimab and cocktails of LY-CoV016 + LY-CoV555 and REGN10933 + REGN10987.
RT-PCR tests have been in use for a while now for SARS-CoV-2 detection and can very well aid in screening. They use a set of two or more primers for detection. Spike Gene Target Failure(SPGF) whereby mutations in the spike gene (as is the case in Omicron), could possibly affect the efficiency of screening. Fortunately, the second primer in the case works in binding to target sites. Considerable number of testing kits have not made primer details publicly available. More studies need to be done in this regard and would be helpful if primer data of testing kits were made available.
An even more saddening and pressing issue is vaccine equity. Anybody who looks at stats of the global doses administered now, every continent except Africa lights up like a christmas tree. Dr Vinod Scaria, scientist at CSIR-IGIB, Delhi via twitter says” If we are not alarmed, we can as well be ashamed.” Dr Glen O Hara says ”The world should provide support to South Africa and Africa and not discriminate or isolate it! “.
South African leaders have done an amazing job and have set an example in letting the world know of the emergence of the variant and in making all available data public. Rather than imposing travel bans and further isolation, supporting Africa with resources, vaccines and aid is the need of the hour. We neglected Africa once, but it's high time we do something about it now.
Misinformation is worse than a pandemic and Dr Emma Hodcroft, co-developer of the Nextstrain tweeted a gentle reminder to “Interpret carefully”. She says “The total number of sequences for the time period is small and may not be biased.” Data on understanding disease severity, clinical manifestations, antibody neutralisation, immune escape and transmissibility of the variant in its entirety is vague at the present moment. But what we can do now is amping up genomic surveillance and boosting vaccination drives. The best way to protect you and your loved ones is by masking up, taking all the doses of the COVID19 vaccine, and being mindful that with proper and timely actions we can overcome this pandemic.
About the author
Author is a researcher at the CSIR Institute of Genomics and Integrative Biology and opinions expressed are personal.

Comments
Post a Comment